About
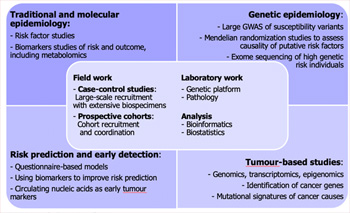
The Genomic Epidemiology Branch (GEM) applies integrative genomic, molecular, and epidemiological approaches to elucidate cancer causes and translate discoveries into prevention. In 2021–2025, GEM pioneered the development of genomic epidemiology, consolidating IARC’s leadership in Understanding the Causes of Cancer by uniting germline, somatic, and exposure data across large international studies and consortia, aligned with the IARC Medium-Term Strategy pillars and emerging priorities.
A flagship achievement is the Mutographs project, which defined distinct mutational signatures across kidney, colorectal, and head and neck cancers – exposing hidden etiological processes and implicating environmental and lifestyle factors, while also revealing signatures of unknown origin. In parallel, GEM expanded discovery of germline susceptibility, advanced multi-omic tumour analyses that refine classification, and progressed early-detection biomarkers, including circulating assays and urinary TERT promoter mutation tests moving towards clinical validation. These outputs were enabled by Open Science infrastructure (IARC Analytical Hub) and GEM’s convening of global networks.
In the strategy for 2026–2030, building on these foundations, GEM’s three-axis programme will: discover exposures (Mutographs-eoCRC, Mutographs-ENV, DISCERN); decode carcinogenesis (PROMINENT, genetic susceptibility consortia, GENESIS, Ecosystems); and deliver precision early detection (genetic risk stratification, WSI-AI, UbioBca, IMPACT-HNC), linking molecular mechanisms to population risk and generating transportable, policy-relevant evidence for prevention. GEM is exceptionally well poised to capitalize on the emerging field of genomic epidemiology – leveraging its consortia leadership, harmonized data assets, and integrated laboratory–compute capabilities to move from association to attribution and mechanistic insight in cancer etiology. With exceptional multidisciplinary expertise, sustained extramural support, and a commitment to training and capacity-building, GEM generates policy-relevant, globally impactful science that advances precision prevention and reinforces IARC’s mandate to reduce the global cancer burden.
Flagship example: Applying genomic epidemiology to cancer causation
A flagship example of the genomic epidemiology approach of GEM is the Mutographs project, a Cancer Research UK Grand Challenge initiative that applies the concept of mutational signatures: characteristic patterns of DNA mutations in tumours that can reveal specific causal exposures.
Within the initial phase of this project, IARC targeted six cancer sites selected for their striking geographical variation in incidence, as identified through IARC’s global cancer statistics. Leveraging IARC’s convening power and extensive experience in international recruitment, GEM coordinated patient enrolment across multiple continents, implementing one of the largest cancer genomics studies of its kind: resulting in whole-genome sequencing of tumour–normal DNA pairs from 5000 patients with cancer.
To ensure scientific rigour and comparability, all recruitment followed a harmonized protocol for biospecimen collection and contextualized epidemiological data capture, tailored to each study setting. GEM oversaw the full pathway from fieldwork to data analyses, including centralized pathology review, high-quality DNA extraction, and quality control of biological samples. Genomic analyses were undertaken with world-leading partners, including the Wellcome Sanger Institute and the University of California, San Diego, applying cutting-edge mutational signature analysis to whole-genome sequencing data. The second phase of this approach (from 2026 onwards) builds on these strengths to answer two distinct questions: (i) what are the causes of early-onset colorectal cancers? and (ii) what are the causes of differences in renal cancer incidence across countries?
This project exemplifies GEM’s integration of:
- Observational epidemiology – identifying cancers, populations, and regions of interest, and embedding detailed behavioural, exposure, and clinical assessment,
- Global collaborative networks – engaging local partners for research contributions, recruitment, and follow-up,
- Operational expertise – designing and implementing complex, multi-country study protocols for sample and data collection,
- Genomics and bioinformatics – applying advanced analytical pipelines to large-scale sequencing data, and
- Data integration – linking molecular patterns to differences in incidence and plausible causal exposures to allow etiological inference.